The characterization, pathotype distribution and genetic diversity of Xanthomonas oryzae pv. oryzae in rice production centers of the Special Region of Yogyakarta, Indonesia
Main Article Content
Abstract
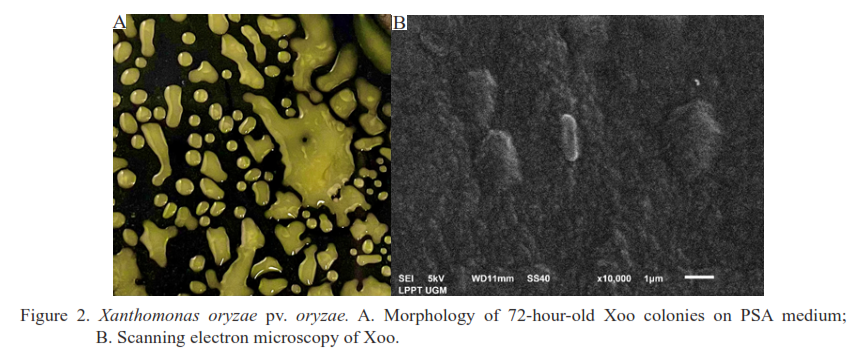
Xanthomonas oryzae pv. oryzae (Xoo) is a pathogenic bacterium that induces bacterial leaf blight in rice. This disease is challenging to treat due to the presence of multiple pathotypes that can harm the plants. Characterizing and determining the distribution of pathotypes and genetic diversity of Xoo in the Special Region of Yogyakarta is essential for evaluating the appropriate approach to managing rice leaf blight in different districts of Yogyakarta. This study aimed to ascertain the characterization, distribution of pathotypes, and genetic diversity of Xoo in the Special Region of Yogyakarta. This study involved the isolation and characterization of Xoo from various rice production centers in Yogyakarta. The distribution of Xoo pathotypes was determined using five differential varieties (Tetep (4251), PB 5 (4827), Java 14 (11022), Kencana Bali (4477), and Kuntulan (1529)). Pathogenicity testing was conducted on six common varieties used by Yogyakarta farmers (IR64, Ciherang, C4, Mekongga, Menthik Wangi, and Inpari 42). Additionally, molecular characterization of Xoo was performed. The bacterial leaf blight that affects rice plants in the Yogyakarta region, caused by Xoo, is identified by yellow circular colonies. It exhibits a negative Gram staining response, positive catalase activity, negative oxidase activity, and does not hydrolyse starch. The Xoo pathotypes identified in Yogyakarta are IV, VIII, and XI. All six prevalent cultivars utilized by farmers in Yogyakarta are susceptible to Xoo. Out of each pathotype, four isolates were chosen, and they were divided into two distinct groups based on the DNA banding pattern they formed. Among these isolates, three had the lowest base sequence at 200 bp, while one isolate had a different DNA banding pattern with the lowest base sequence between 250–300 bp.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
References
Adachi N, Tsukamoto S, Inoue Y, & Azegumi K. 2012. Control of bacterial seedling rot and seedling blight of rice by bacteriophage. Plant Dis. 96(7) 1033–1036. https://doi.org/10.1094/pdis-03-11-0232-re
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, & Struhl, K. 2003. Current Protocols in Molecular Biology. John Wiley & Sons, Inc. New Jersey.
Cappucino JG & Sherman N. 2001. Microbiology: A Laboratory Manual. Second edition. Benjamin Cummings Publishing Company. New York.
Clarridge JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17(4): 840–862. https://doi.org/10.1128%2FCMR.17.4.840-862.2004
Djatmiko HA, Prakoso B, & Prihatiningsih N. 2011. Penentuan patotipe dan keragaman genetik Xanthomonas oryzae pv. oryzae pada tanaman padi di wilayah Karesidenan Banyumas [Identification of pathotype and genetic diversity of Xanthomonas oryzae pv. oryzae on rice in Karesidenan Banyumas area]. J. Trop. Plant Pests Dis. 11(1): 35–46. https://doi.org/10.23960/j.hptt.11135-46
EPPO. 2007. Xanthomonas oryzae. EPPO Bulletin. 37(3): 543–553. https://doi.org/10.1111/j.1365-2338.2007.01162.x
Ghasemie E, Kazempour MN, & Padasht F. 2008. Isolation and identification of Xanthomonas oryzae pv. oryaae the causal agent of bacterial blight of rice in Iran. J. Plant Prot Res. 48(1): 53–62. https://doi.org/10.2478/v10045-008-0006-9
Islam MdR, Alam MdS, Khan AI, Hossain I, Adam LR, & Daayf F. 2016. Analyses of genetic diversity of bacterial blight pathogen, Xanthomonas oryzae pv. oryzae using IS1112 in Bangladesh. C. R. Biol. 399(9–10): 399–407. https://doi.org/10.1016/j.crvi.2016.06.002
Lang JM, Hamilton JP, Diaz MGQ, Van Sluys MA, Burgos MRG, Cruz CMV, Buell CR, Tisserat NA, & Leach JE. 2010. Genomics-based diagnostic marker development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis. 94(3): 311–319. https://doi.org/10.1094/pdis-94-3-0311
Lee KS, Rasabandith S, Angeles ER. & Khush GS. 2003. Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathology. 93(2): 147–152. https://doi.org/10.1094/phyto.2003.93.2.147
Niño-Liu DO, Ronald PC, & Bogdanove AJ. 2006. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 7(5): 303–324. https://doi.org/10.1111/j.1364-3703.2006.00344.x
Müller CMO, Yamashita F, & Laurindo JB. 2008. Evaluation of the effects of glycerol and sorbitol concentration and water activity on the water barrier properties of cassava starch films through a solubility approach. Carbohydr. Polym. 72(1): 82–87. https://doi.org/10.1016/j.carbpol.2007.07.026
Ranjani P, Gowthami Y, Samuel, Gnanamanickam S, & Palani P. 2018. Bacteriophages: A new weapon for the control of bacterial blight disease in rice caused by Xanthomonas oryzae. Microbiol. Biotechnol. Lett. 46(4): 346–359. https://doi.org/10.4014/mbl.1807.07009
Sudir, Suprihanto, & Kadir TS. 2009. Identifikasi patotipe Xanthomonas oryzae pv. oryzae penyebab penyakit bercak daun bakteri pada daerah sentra produksi padi di Jawa [Pathotype identification of Xanthomonas oryzae pv. oryzae, causing bacterial leaf blight in rice production centers in Java]. Jurnal Penelitian Pertanian Tanaman Pangan. 28(3): 131–138.
Sudir, Yogi YA, & Syahri. 2013. Komposisi dan sebaran patotipe Xanthomonas oryzae pv. oryzae di sentra produksi padi di Sumatera Selatan [Composition and distribution pathotypes of Xanthomonas oryzae pv. oryzae in the rice production center in South Sumatra]. Penelitian Pertanian Tanaman Pangan. 32(2): 98–108.
Sudir & Yuliani D. 2016. Composition and distribution of Xanthomonas oryzae pv. oryzae pathotypes, the pathogen of rice bacterial leaf blight in Indonesia. AGRIVITA Journal of Agricultural Science. 38(2): 174–185. https://dx.doi.org/10.17503/agrivita.v38i2.588
Suparyono, Sudir, & Suprihanto. 2004. Pathotype profile of Xanthomonas oryzae pv. oryzae isolates from the rice ecosystem in Java. Agrivita Journal of Agriculture Science. 5(2): 63–69.
Suprihatno B, Darajat AA, Satoto, Baehaki SE, Suprihanto, Setyono A, Indrasari SD, Wardana IP, & Sembiring H. 2010. Deskripsi Varietas Padi: Way Apo Buru [Rice Variety Description: Way Apo Buru]. Balai Besar Penelitian Tanaman Padi. Departemen Pertanian RI. Jakarta.
Tian Y, Zhao Y, Xu R, Liu F, Hu B, & Walcott RR. 2014. Simultaneous detection of Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola in rice seed using a padlock probe-based assay. Phytopathology. 104(10): 1130–1137. https://doi.org/10.1094/phyto-10-13-0274-r
Wahyudi NA. 2021. BPS sebut potensi lahan panen padi tahun ini menyusut 0,14 juta hektar [BPS says the potential area for rice harvesting will shrink by 0.14 million this year]. https://m.bisnis.com/amp/read/20211015/12/1454724/bps-sebut-potensi-luas-lahan-panenpadi-menyusut-014-juta-tahun-ini.Accessed 27 January 2022.
Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan J, Yang S, Hu L, Leung H, Mew TW, Teng PS, Wang Z, & Mundt CC. 2000. Genetic diversity and disease control in rice. Nature. 406: 718– 722. https://doi.org/10.1038/35021046
Yashitola J, Krishnaveni D, Reddy APK, & Sonti RV. 1997. Genetic diversity within the population of Xanthomonas oryzae pv. oryzae in India. Phytopathology. 87(7): 760–765. https://doi.org/10.1094/phyto.1997.87.7.760